This is a talk on some of our recent work on discovering genetic variants associated with human cortical structure and how those variants influence gene regulatory potential in the developing brain. The talk was given on April 14, 2020 as a virtual seminar on Zoom due to the COVID crisis.
Dan Liang wins Dissertation Completion Fellowship
Congratulations to Dan Liang, who won the Dissertation Completion Fellowship! She’ll be finishing up her dissertation work this year on chromatin accessibility differences during human cortical differentiation.
Welcome to Felix Kyere, new neuroscience graduate student
We’re happy to have Felix Kyere, a graduate student in the neuroscience graduate program, join our lab for his dissertation work. Welcome Felix!
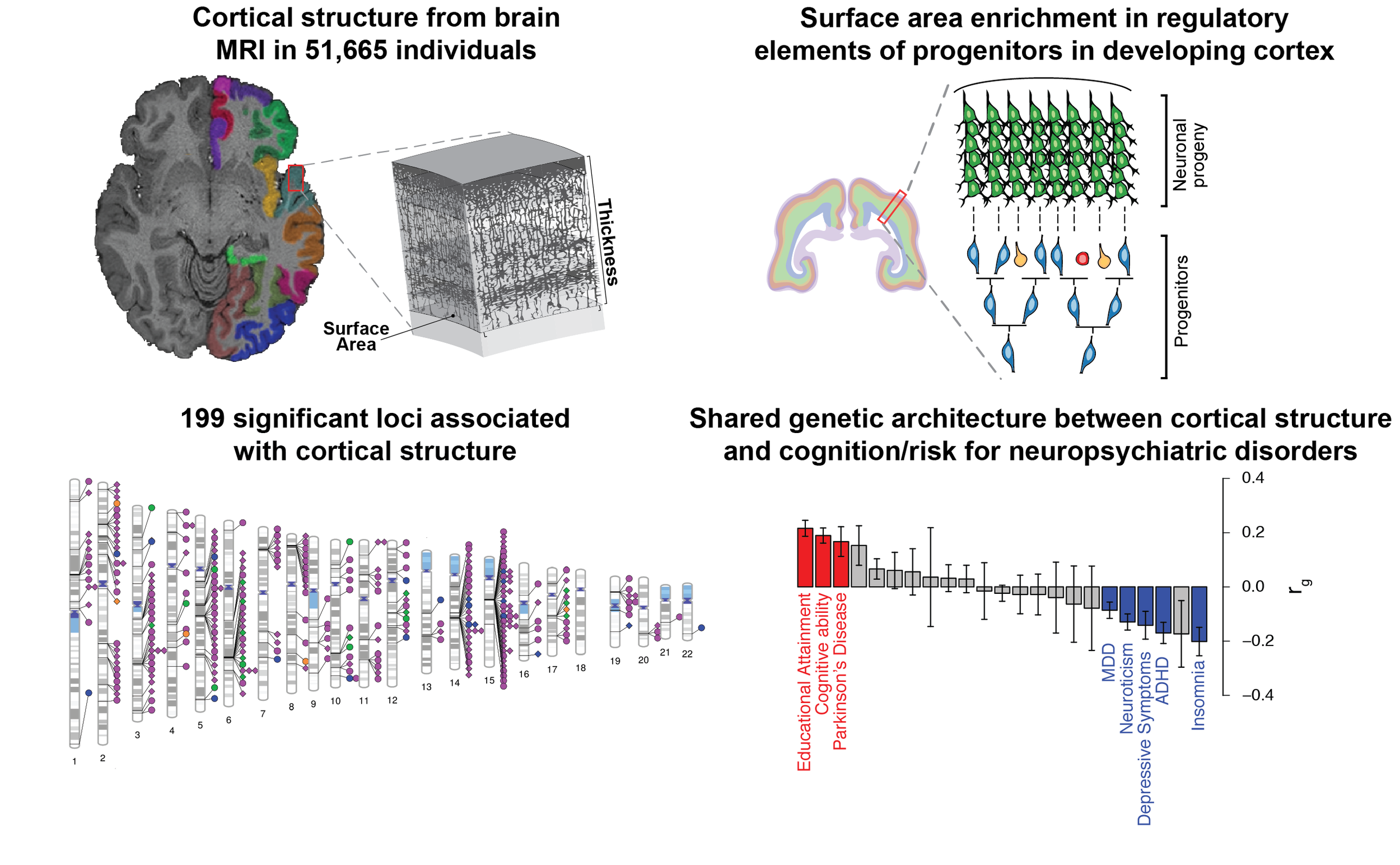
GWAS of Cortical Structure published in Science
As part of the ENIGMA consortium, we published a large study to demonstrate how genetic variation in humans impacts cortical structure. We identified 199 loci that were significantly associated with cortical structure by a stringent criterion. We also found that those genetic variants that influence adult surface area are enriched in genomic regions active in progenitor cells during neuronal differentiation well before birth. This is evidence in humans supporting the Radial Unit Hypothesis, put forward by Pasko Rakic in the 1980s. In addition, there were genetic correlations detected between cortical surface area and several traits including cognitive abilities and ADHD. This implies that genetic variants that impact cortical structure also impact cortical function. You can find our work published here. This is a huge collaboration and wouldn’t be possible without the joint work of literally hundreds of scientists. We started the ENIGMA consortium about 10 years ago, and this is our latest and greatest work. It’s particularly exciting because the structure of the cortex allows us to have human specific abilities and disruption of cortical structure is thought to cause neuropsychiatric disorders.
Pre-print posted on genome-wide association study in autism using the SPARK dataset
In joint work with the lab of Hyejung Won by joint postdoc Nana Matoba, we used the SPARK dataset to identify common genetic variants associated with risk for Autism Spectrum Disorder. We were able to replicate previous findings and also identify a new locus. For that new locus, we used a multiplex parallel reporter assay (MPRA) to identify a potential causal variant. We integrated this information with existing eQTL resources to identify genes of action for those variants. You can read more about this work in the pre-print here.
Biorxiv pre-print posted on genetic effects on chromatin accessibility during human neuronal differentiation
Dan Liang, a graduate student in the lab, posted a pre-print of our work on genetic effects on chromatin accessibility in cultured human neural progenitor cells. In this work, we identified thousands of common genetic variants that influence gene regulatory activity during human neuronal differentiation. We use this data to explain the gene regulatory mechanisms of several GWAS loci for neuropsychiatric disorders and brain relevant traits. The preprint can be found here.
Dan Liang wins a travel award to attend the World Congress of Psychiatric Genetics
Bioinformatics and Computational Biology graduate student won a travel award to attend the World Congress of Psychiatric Genetics meeting in Anaheim, CA. Her talk is entitled: “Genetic Variants Affecting Chromatin Accessibility During Human Neuronal Differentiation”.
Biorxiv pre-print posted on evolutionary genomics of human brain Structure
Together with Simon Fisher and Amanda Tilot, we just submitted a biorxiv pre-print on how genetic variation through our evolutionary history impacted modern human brain structure. Interesting findings include that variation present within human-specific regulatory elements present in the developing cortex, prior to birth, have a strong impact on adult brain structure. And, alleles with evidence of selective pressure over very recent time scales (in evolutionary history) have impacts on specific brain regions, including those involved in spoken language and visual processing. Read more about this work here: https://www.biorxiv.org/content/10.1101/703793v1.
Mike Lafferty selected to attend Human Genomics Summer School.
Michael Lafferty, BCB Graduate Student in our lab, has been selected to attend the Leena Peltonen School of Human Genomics program in Switzerland on August 18-22, 2019.
Brandon Le wins training grant
Congratulations to Brandon Le who won the Bioinformatics and Computational Biology T32 training grant!
Undergraduate Tianyi Liu meets the UNC Chancellor at Poster Presentation
Tianyi Liu presented his research at the Celebration of Undergraduate Research and got to meet the Chancellor of UNC! Tianyi looks very happy :)
The Stein Lab welcomes 2 new Bioinformatics and Computational Biology Graduate Students
We’re very pleased to welcome Brandon Le and Nil Aygun to our lab! Both are students in the Bioinformatics and Computational Biology graduate program and will work with us for their dissertations. Both also have both human neural stem cell culture and bioinformatics experience as well. Excited to have them on board! See more about them here.
SPARK grant acquired to identify common variants associated with autism
Our lab and the lab of Hyejung Won (wonlab.org) acquired a grant from the Simons Foundation to identify common genetic variants associated with risk for autism. Our shared postdoc Nana Matoba is working on this project. Thanks to the Simons Foundation for their support and we hope to add to the list of common genetic variants creating risk for autism.
Review article published on imaging genetics
Brandon Le (grad student in Bioinformatics and Computational Biology) and I wrote a review on how imaging genetics can fit as one important piece in understanding mechanisms by which genetic variants create risk for neuropsychiatric disorders. We also detail what we think are some important next steps for imaging genetics research. See the paper here: https://www.ncbi.nlm.nih.gov/pubmed/30864184
Grad students win awards at the UNC Genetics Retreat in Wilmington
Dan Liang was selected to give a talk on the genetic influences of chromatin accessibility during human neuronal differentiation at the UNC genetics retreat in Wilmington, and won the best student talk of the Bioinformatics and Computational Biology program. Congrats Dan! Mike Lafferty presented a poster on miRNAs involved in human neurogenesis and won a best student poster award for the Bioinformatics and Computational Biology program. Congrats Mike!
Moving to Mary Ellen Jones Building at UNC
We are moving on March 19th, along with the UNC Neuroscience Center, to the newly renovated, window filled, Mary Ellen Jones Building! Looks really beautiful and will be a great place to do some genetics and brain science.
Undergraduates in the Stein lab off to bigger and better things!
Congratulations to Tianyi Liu, who joined our lab in 2017 and just was admitted to the UNC Biostatistics PhD program.
Also, congratulations to Leo Zsembik, the first undergraduate in the Stein lab, who was just admitted to the Oxford Neuroscience Master’s program.
We wish you both best of luck in your next scientific adventures!
Teaching middle schoolers about the brain
Today we taught some middle school students about how genetics shapes the brain, and let them explore a human brain in virtual reality.
Andrew S. Rachlin UNC Neuroscience Symposium
The Stein lab co-organized the Andrew S. Rachlin UNC Neuroscience Symposium this year. Thanks to Dan Geschwind and Flora Vaccarino for coming as our keynote speakers, and to all the local speakers for a fascinating seminar! Hope it leads to future collaborations and projects on tackling neuropsychiatric disorder genetics here in the triangle area.
Stein lab at the Walk for Hope
We ran the 5k (Jason) and 10k (Tianyi) at the Walk for Hope to raise money for the Foundation of Hope. This is a great organization supporting research into mental health, including supporting some of our work. https://walkforhope.com/