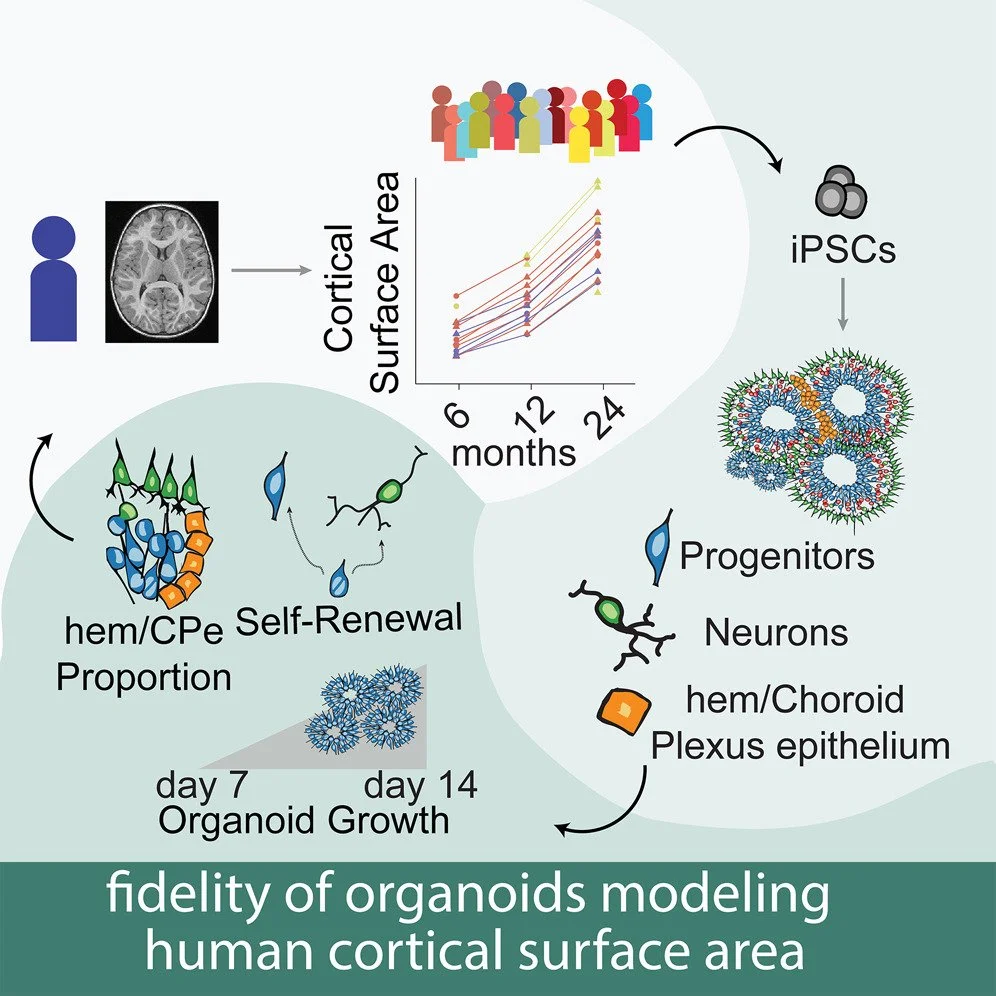
We published our first paper as part of the Infant Brain Imaging Study (IBIS) induced pluripotent stem cell (iPSC) project in Cell Stem Cell. We generated iPSCs from 18 participants in the infant brain imaging study who had longitudinal brain structural images acquired since infancy. Features from cortical organoids correlated with the brain growth of the individual from which they were derived. For example, gene expression at the crux of fate decisions between neural progenitor self renewal and neuronal differentiation were strongly associated with cortical surface area. Congrats to Rose Glass, Nana Matoba and all the wet, dry, and clinical labs that made such an ambitious project possible!
Jordan Valone Defends his Dissertation
Dr. Jordan Valone successfully defended his dissertation in Bioinformatics and Computational Biology! Jordan co-led our work on context-dependent genetic regulation and pharmacogenomics. Congratulations, Jordan!



Stein Lab at UNC Neuroscience Retreat
At the UNC Neuroscience Retreat, Jason gave an impromptu lecture on the interaction between bees and our most inspirational scientist, Dr. Nicholas Cage. And, he was more than honored to receive the “Mentor of the Year” award from the UNC Neuroscience Curriculum!



Stein lab at ASHG 2025
We attended the American Society of Human Genetics in Boston, where we had a lab reunion dinner! It was amazing to see everyone again.


Clear IT! Tissue Clearing Workshop at UNC
Together with colleagues from our sister university, the University of Tübingen in Germany, we organized a tissue clearing conference bringing experts from around the triangle to discuss new research in 3D image acquisition and analysis in April, 2025.
Preprint on molecular gene x treatment effects
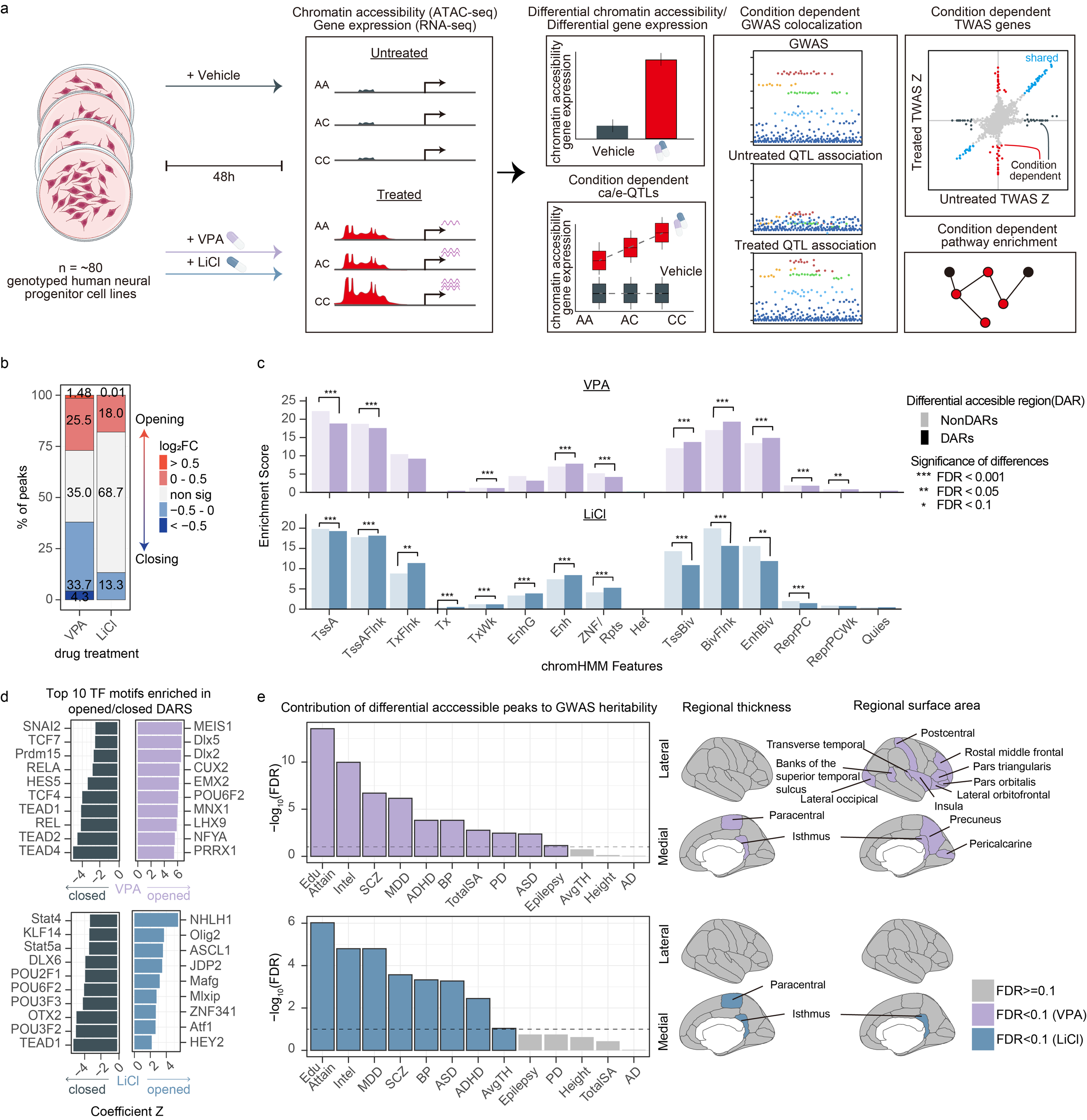
We released a preprint on VPA and Li molecular quantitative trait loci in primary human neural progenitors. Gene by treatment (GxT) interactions are critical for understanding psychiatric disorder risk and variability in treatment response, yet their discovery remains challenging due to the complexity of human exposures. For example, while epidemiological birth cohorts demonstrated that valproic acid (VPA) taken during pregnancy strongly increases the likelihood of an autism spectrum disorder (ASD) diagnosis in the exposed child, only a small proportion of those exposed are affected with ASD. Similarly, while lithium has been successfully used as a treatment for bipolar disorder for over 75 years, around half of individuals treated have a beneficial clinical response. It is likely that genetic variation plays a strong role in response to each treatment, but pharmacogenomic studies are often underpowered and confounded by poorly controlled concentration, duration, and adherence of treatment. Here, we demonstrate a novel approach to dissect these interactions by investigating how common genetic variation alters gene regulatory responses to clinically relevant perturbations, which we call “GxT in a dish”, where the molecular response of each donor to well-controlled exposures can be precisely measured.
Jason is named a Yang Family Biomedical Scholar
Jason was named a Yang Family Biomedical Scholar by the UNC School of Medicine. Thank you to the Stein lab team over the years who have done such an amazing job discovering how genetics change human brain development and function that enabled this recognition. Thank you to the Department of Genetics and School of Medicine for nominating me. And, thank you to the Yang Family for funding our work! https://news.unchealthcare.org/2025/04/unc-school-of-medicine-names-2024-25-yang-family-biomedical-scholars/
Maya Yin joins the lab
We welcome Maya Yin, a graduate student in the Bioinformatics and Computational Biology program, to the lab!
Developmental Diversity in a Dish talk
You can watch a seminar on our recent work presented at the Developmental Diversity in a Dish seminar series, found here: https://www.youtube.com/playlist?list=PLbeFhDgIkXxd-3CLdfuWvGguZZOveIso4.
Continued domination at chili cookoff
For the second year in a row, the Stein lab has placed in the UNC Neuroscience Chili Cookoff. Jason placed 3rd as assessed by Chili Reviewers 1-3, and Ariana and Alvaro placed second in the People’s Choice Award. Here’s to some tasty chili.
Preprint on regulatory activity of cortical structure associated variants
We released a preprint on the regulatory activity of cortical structure associated variants. We conducted a multiplexed parallel reporter assay using common variants associated with inter-individual differences in the size of the human cortex. Most loci had a least one variant with regulatory potential, and a small subset of these loci showed Wnt-dependent activity. Those loci with regulatory activity were strongly enriched in Alu elements, a type of retrotransposon that has increased in prevalence along the human lineage together with brain size increases. These results help to explain the molecular mechanisms leading to differences in the structure of the human cortex. You can read more about our study here.
Preprint on organoid modeling of brain growth
We released a preprint describing organoid modeling of brain growth. Through a collaboration with the Infant Brain Imaging Study, we generated induced pluripotent stem cells (iPSCs) from 18 participants who had undergone longitudinal brain imaging during infancy. We differentiated these iPSCs into cortical organoids and studied how cellular and molecular processes occurring in these brain models in a dish recapitulate the brain growth of the individual from which they were derived. We found that gene expression levels within progenitor cells at the crux of fate decisions - deciding to become a neuron or another progenitor - were critical for determining inter individual differences in brain growth. These findings support the fidelity of cortical organoids as a model system. You can read more about our work here.
Context-dependent eQTL/caQTL paper published in Nature Neuroscience
Gene regulatory effects have been difficult to detect at many non-coding loci associated with brain-related traits, likely because some genetic variants have distinct functions in specific contexts. To explore context-dependent gene regulation, we measured chromatin accessibility and gene expression after activation of the canonical Wnt pathway in primary human neural progenitors. We identified many context-dependent genetic effects, some of which help explain the mechanisms underlying brain-related traits. This work was published in Nature Neuroscience.
IDDRC organoid cross-site reproducibility project published in Stem Cell Reports
We evaluated the level of reproducibility of organoid differentiation using the same iPSC line differentiated multiple times across 3 different sites. Cell type proportions and cortical wall organization were reproducible, but metabolic gene expression levels as well as size did exhibit variability across sites. We hope this will inspire cross-site meta-analyses to increase sample sizes in iPSC-derived organoid studies. You can read more about our work here.
Genetics of cell-type-specific post-transcriptional gene regulation during human neurogenesis published in AJHG
There are many types of gene regulation occurring in the human genome that tune not just the level of expression of a gene, but also RNA editing and alternative polyadenylation. These forms of gene regulation are poorly studied during human development. Here, we show that RNA editing and polyadenylation are highly cell type specific, and that genetic variation impacts these post-transcriptional modifications. Our work was published in the American Journal of Human Genetics.
Stein lab at the UNC Neuroscience Retreat
Sporting the latest in neuroscience fashion, the Stein lab joins the UNC Neuroscience Retreat!
Karthik Eswar successfully defends his undergraduate honors thesis
Karthik Eswar, an undergraduate student majoring in Computer Science and Neuroscience, successfully defended his undegraduate honors thesis entitled “Convolutional Neural Network based Enhancement and Restoration of Mouse Brain Light Sheet Images”. Congrats Karthik!
The Stein lab wins second place in the Chili Cookoff
At the inaugural UNC Neuroscience Center Chili Cookoff, the Stein lab won a surprising second place! As always, keeping our science and our chili spicy.
Newsletter on organoid creation for IBIS iPSC participants
Please see our newsletter to the IBIS iPSC participants, shown below
Rubal Singla wins Autism Science Foundation Fellowship
Congrats to Rubal Singla who won the Autism Science Foundation Fellowship. Her grant application is entitled “Can Brain Size Predict Autism? Building a Model System with iPSC-derived Cortical Organoids”. You can read more about her grant here.